Mycoplasma pneumoniae
The minimal gene set to sustain life
Large-scale quantitative analyses of Mycoplasma pneumoniae
People involved: Eva Yus, Maria Lluch, Sira Martínez, Javier Delgado, Marie Trussart, Besray Unal, Tony Ferrar, Verónica Llorens and Carolina Gallo
A great effort has been made in the past years towards the identification of the minimal gene complement necessary for life, especially with the boom of synthetic biology. In fact the task has been driven by the considerable sequencing projects in place, particularly in the prokaryote kingdom.
Such computation analysis has been very powerful, however comparative genomics, aiming at finding the common gene set among bacteria, can lead to underestimations given the fact that different organisms can use different genes to perform the same function.
Furthermore, forward genetics approaches have provided us with lists of essential genes that are believed to represent the basic core for a living cell, whereas the rest of the genes, though apparently dispensable, must be there for a reason. Intriguingly many of those essential genes revealed to have unknown functions (albeit been conserved in evolution), functions that remain obscure after this high-throughput analysis. This highlights how far we are of unveiling the biology of even “simple” unicellular organisms.
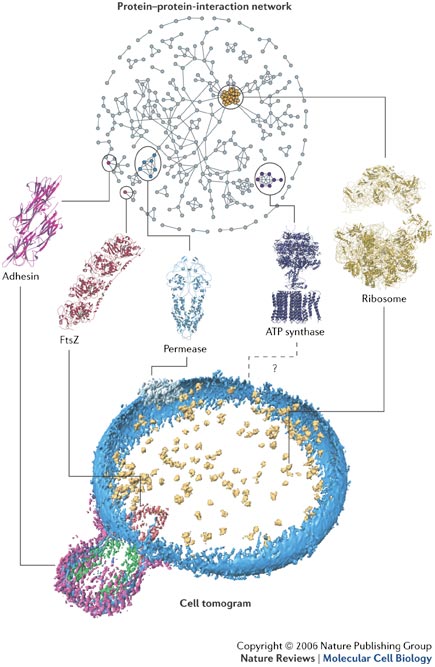
The philosophy in the lab is that in order to understand the basics of a living device, we must go further and undertake a global analysis at every level: from genomics to metabolomics, including gene expression profiling, proteome assessment, survey of the protein-protein interactions and so on. Such approaches aiming at the whole quantitative understanding of an entire organism for the first time are expected to guide rational engineering/modifications.
For this goal we have chosen Mycoplasma pneumoniae as model organism. Mycoplasma sp. are the smallest self-replicating organisms (~500-1000 genes). For this reason they are suitable models for systems biology and are thought to fit the minimal genome paradigm. They are parasites depending on their hosts, and their metabolism is scarce, reflecting the strong genomic simplification they have suffered from a bacillus-like ancestor. Nevertheless, they can be maintained in vitro.
The Mycoplasma consortium
This project started at the European Molecular Biology Laboratory on 2006: a big team of researchers (inspired by the father of Mycoplasma pneumoniae -omics, Porf. Richard Herrmann at Heidelberg University) joined to develop genetic and molecular biology tools, decipher the regulation of gene expression, the metabolic fluxes and accomplish the modelling of Mycoplasma pneumoniae physiology in the framework of the SmartCell cell simulator. Our lab did a big part of the experimental effort (metabolomics, proteomics, transcriptomics, genomics...), Peer Bork and colleagues did the bioinformatics and data integration. Rob Russell lab worked on bioinformatics of protein complexes. Anne-Claude Gavin group was responsible for the interactome. Achilleas Frangakis shares his expertise on cryoelectron tomography
Regulation of gene expression
People involved: Eva Yus, Besray Unal, Veronica Llorens and Marie Trussart
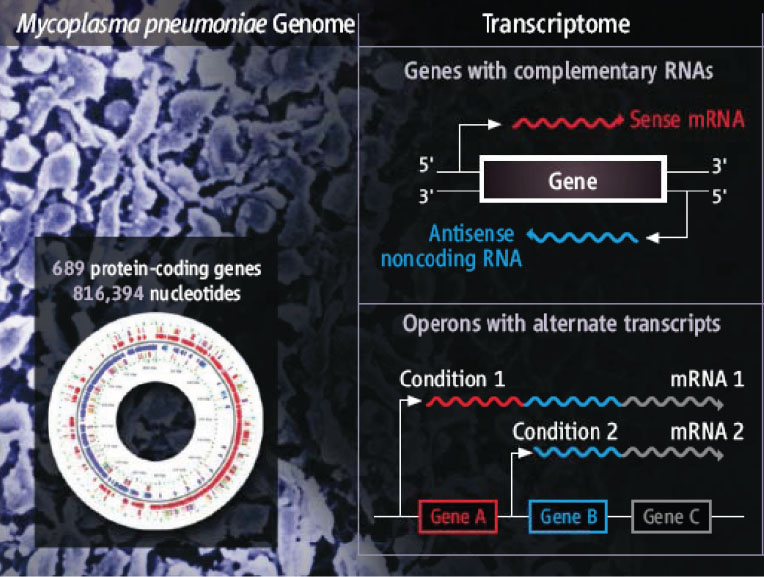
Transcriptome-wide studies have been wide-spread and highly effective in eukaryotes for more than a decade. However the transcriptomes of bacteria and archaea have been largely overlooked until recently.
A recent study by our lab revealed impressive transcriptome complexity. An unexpected abundance of antisense transcription and alternative transcripts within operons suggested a highly structured multifaceted regulatory machinery. Yet, this organism has few transcription factors encoded in its genome, suggesting additional layers of regulation.
At these point we realized we needed to get more insight on transcriptcional regulation, with these main goals:
- Study RNA processing and half-life.
- Identify all putative transcription factors and binding sites.
- Perform reverse engineering of the gene regulatory network based on our large expression dataset.
- Obtain a 3D model of M. pneumoniae chromatin in order to find out the role in transcriptcional regulation.
Modeling a whole cell
People involved: Veronica Llorens
Probably there is not so much data collected for one single organism as there is for Mycoplasma pneumoniae. Recently, a paper was published on a whole cell model on its close relative Mycoplasma genitalium.
We aim at implementing this model in M. pneumoniae and improve it by adding new data obtained in our lab (see former section).
Bacterial cell division
People involved: Maria Lluch
The aim of this project is study the bacterial cell divison using as models Mycoplasma pneumoniae and E. coli. While the cell division operon in E.coli comprises 14 genes in M. pneumoniae it comprises only four genes (mraZ, mraW, ftsA-like and ftsZ). The objective of this work is to elucidate the connection between these proteins in terms of functionality and decipher the mechanistics of the regulation of bacterial cell division.
Gene essentiality in a minimal bacterium
People involved: Maria Lluch, Judith Wodke* and Javier Delgado
Study the M. pneumoniae gene essentialness in different conditions. For this propose, we have designed new strategies, using new ultra-sequencing technologies.
Developing genetic tools
People involved: Bernhard Pätzold*, Tony Ferrar, Carolina Gallo and Maria Lluch
The aim of this project is to develope a series of genetic tools that are so far restrinting the study and the engeneeing of M. pneumoniae. In particular, we would like to set up self replicating plasmids, create inducible promoters, or restore the double recombination.
Comparative study of M. genitalium and M. pneumoniae
People involved: Maria Lluch
Study the transcriptome, the proteome and the DNA methylome between these two closer related spices to see the conservation at level of antisense transcription and regulatory sequences. The aim of this project is study the impact of DNA methylation in the regulation of transcription and the conservation between species.
* Old lab members